|
||||||||
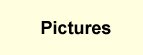 |
 |
 |
 |
 |
| Conformational Analysis of an Antineoplastic Class of Macrocycles | ||
|
Recent studies on the bio-potency of Sansalvamide
A derivatives show promising properties against pancreatic, colon,
breast, prostate, and melanoma cancers. Recent NCI panels have also
proven potency on leukemia, thus a more detailed analysis of the
structural features within the macrocycles is needed to explore
further roles of the molecules. Sansalvamide A (San A) is a marine
fungal product that was discovered by William Fenical. Through structural
manipulation of peptide derivatives based on structure-activity
relationship (SAR) and 2-D NMR, valuable trends arise to provide
a systematic means of controlling bio-potency. Through the use of
various biological assays as well as computational resources it
is possible to analyze the effect of such conformational changes
and gain insight into the development of more potent compounds.
| ||
Multiple derivatives of the San A scaffold were synthesized
and their activity on a pancreatic cancer cell line PL-45 and the
colon cancer cell line HCT-116 is reported here. Then, using MacroModel
within Maestro, we have validated the preferred conformation of many
San A derivatives. Using Monte Carlo methods as well as conformational
constraint and limiting electrostatic and steric qualifications, a
pool of conformers was created. By arranging the lowest energy conformer
within NAMFIS and using 2D NMR experiments NOESY and ROESY predictive
studies of the proposed protein target of the derivatives are described.
By performing pull-down assays we have determined that the protein
target of these derivatives is Hsp90, and therefore computational
validation combined with incorporation of the co-crystal structure
of Hsp90-drug can be used to determine the active conformation of
the molecule when bound to Hsp90. This crystalline matrix will then
be analyzed using NAMFIS, a NMR based modeling program, to create
a dynamical picture of the Sansalvamide A derivatives. Future potent
derivatives can then be predicted using this hybrid computational/experimental
approach. |
||
| • Other Abstracts • | ||